Load Packages
library(tidyverse)
library(moderndive) #install before loading
library(infer) #install before loading
library(fivethirtyeight) #install before loading
library(skimr)
What is the average age of members that have served in congress?
set.seed(123)
congress_age_100 <- congress_age %>%
rep_sample_n(size=100)
1.) Construct the confidence interval
congress_age_100 %>%
specify(response = age)
Response: age (numeric)
# A tibble: 100 x 1
age
<dbl>
1 53.1
2 54.9
3 65.3
4 60.1
5 43.8
6 57.9
7 55.3
8 46
9 42.1
10 37
# ... with 90 more rowscalculate
function (x, stat = c("mean", "median", "sum", "sd", "prop",
"count", "diff in means", "diff in medians", "diff in props",
"Chisq", "F", "slope", "correlation", "t", "z", "ratio of props",
"odds ratio"), order = NULL, ...)
{
check_type(x, tibble::is_tibble)
check_type(stat, rlang::is_string)
check_for_numeric_stat(x, stat)
check_for_factor_stat(x, stat, explanatory_variable(x))
check_args_and_attr(x, explanatory_variable(x), response_variable(x),
stat)
check_point_params(x, stat)
if (!has_response(x)) {
stop_glue("The response variable is not set. Make sure to `specify()` it first.")
}
if (is_nuat(x, "generate") || !attr(x, "generate")) {
if (!is_hypothesized(x)) {
x$replicate <- 1L
}
else if (stat %in% c("mean", "median", "sum", "sd", "prop",
"count", "diff in means", "diff in medians", "diff in props",
"slope", "correlation", "ratio of props", "odds ratio")) {
stop_glue("Theoretical distributions do not exist (or have not been ",
"implemented) for `stat` = \"{stat}\". Are you missing ",
"a `generate()` step?")
}
else if (!(stat %in% c("Chisq", "prop", "count")) & !(stat %in%
c("t", "z") & (attr(x, "theory_type") %in% c("One sample t",
"One sample prop z", "Two sample props z")))) {
return(x)
}
}
if ((stat %in% c("diff in means", "diff in medians", "diff in props",
"ratio of props", "odds ratio")) || (!is_nuat(x, "theory_type") &&
(attr(x, "theory_type") %in% c("Two sample props z",
"Two sample t")))) {
order <- check_order(x, explanatory_variable(x), order)
}
if (!((stat %in% c("diff in means", "diff in medians", "diff in props",
"ratio of props", "odds ratio")) || (!is_nuat(x, "theory_type") &&
attr(x, "theory_type") %in% c("Two sample props z", "Two sample t")))) {
if (!is.null(order)) {
warning_glue("Statistic is not based on a difference or ratio; the `order` argument",
" will be ignored. Check `?calculate` for details.")
}
}
result <- calc_impl(structure(stat, class = gsub(" ", "_",
stat)), x, order, ...)
if ("NULL" %in% class(result)) {
stop_glue("Your choice of `stat` is invalid for the types of variables `specify`ed.")
}
result <- copy_attrs(to = result, from = x)
attr(result, "stat") <- stat
if (nrow(result) == 1) {
result <- select(result, stat)
}
result
}
<bytecode: 0x000000001931f4d8>
<environment: namespace:infer>2.) generate 1000 replicates of your sample of 100
Response: age (numeric)
# A tibble: 100,000 x 2
# Groups: replicate [1,000]
replicate age
<int> <dbl>
1 1 42.1
2 1 71.2
3 1 45.6
4 1 39.6
5 1 56.8
6 1 71.6
7 1 60.5
8 1 56.4
9 1 43.3
10 1 53.1
# ... with 99,990 more rows3.) calculate the mean for each replicate
bootstrap_distribution_mean_age <- congress_age_100 %>%
specify(response = age) %>%
generate(reps = 1000, type = "bootstrap") %>%
calculate(stat = "mean")
bootstrap_distribution_mean_age
# A tibble: 1,000 x 2
replicate stat
* <int> <dbl>
1 1 53.6
2 2 53.2
3 3 52.8
4 4 51.5
5 5 53.0
6 6 54.2
7 7 52.0
8 8 52.8
9 9 53.8
10 10 52.4
# ... with 990 more rows4.) visualize the bootstrap distribution
visualize(bootstrap_distribution_mean_age)
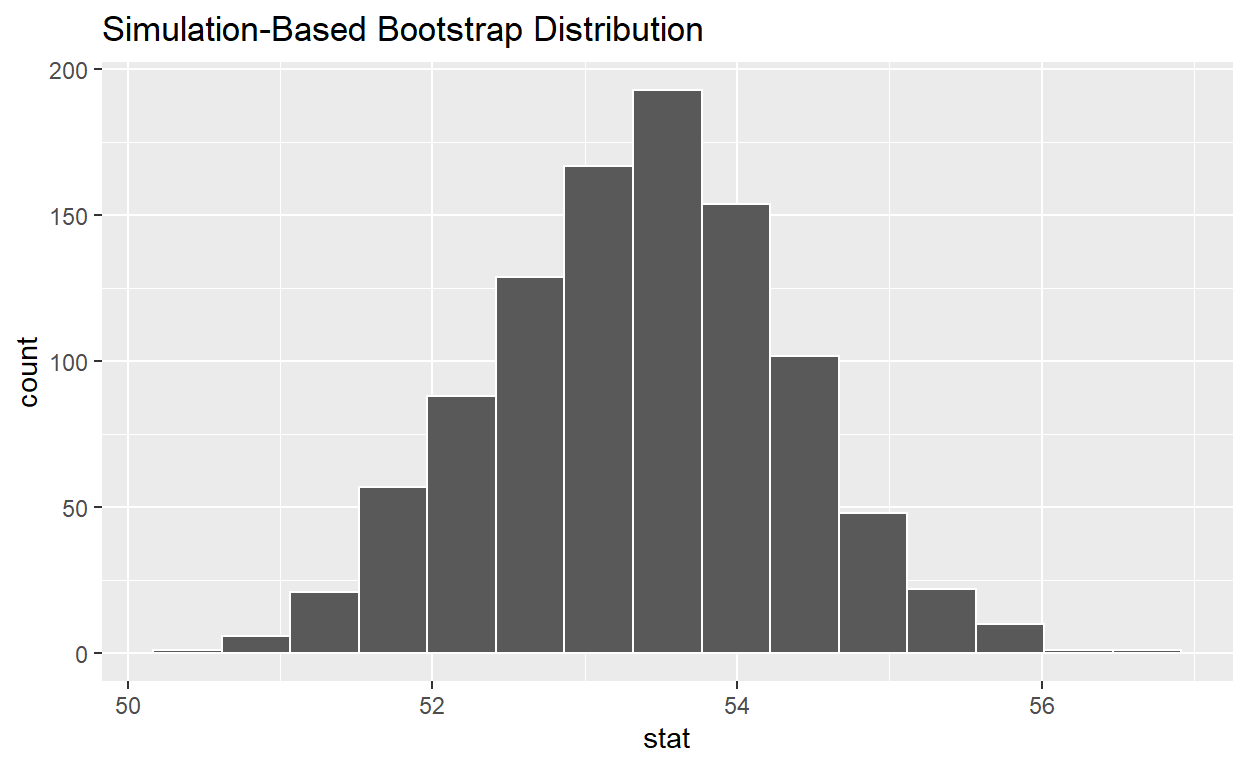
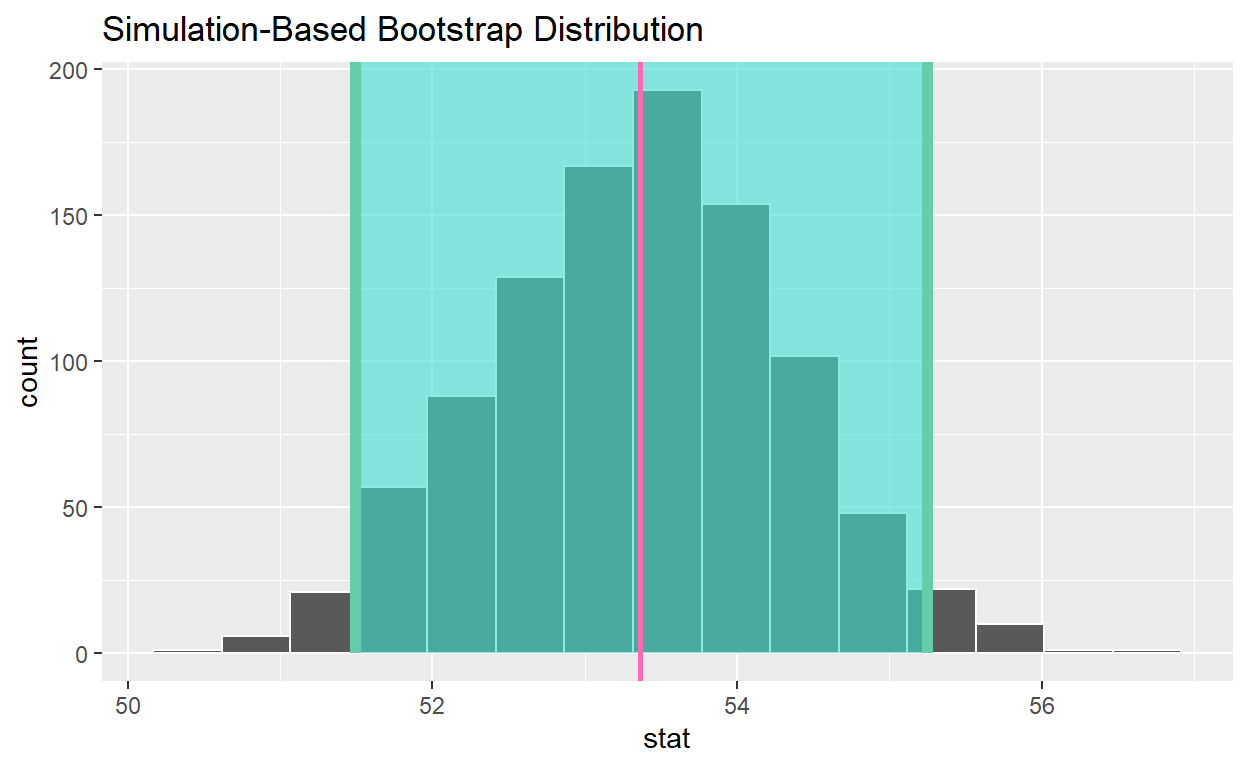
Calculate the 95% confidence interval using the percentile method
congress_ci_percentile <- bootstrap_distribution_mean_age %>%
get_confidence_interval(type = "percentile", level = 0.95)
congress_ci_percentile
# A tibble: 1 x 2
lower_ci upper_ci
<dbl> <dbl>
1 51.5 55.2Calculate the observed point estimate of the mean and assign it to obs_mean_age
obs_mean_age <- congress_age_100 %>%
specify(response = age) %>%
calculate(stat = "mean") %>%
pull()
obs_mean_age
[1] 53.36shade confidence interval
visualize(bootstrap_distribution_mean_age) +
shade_confidence_interval(endpoints = congress_ci_percentile ) +
geom_vline(xintercept = obs_mean_age, color = "hotpink", size = 1 )
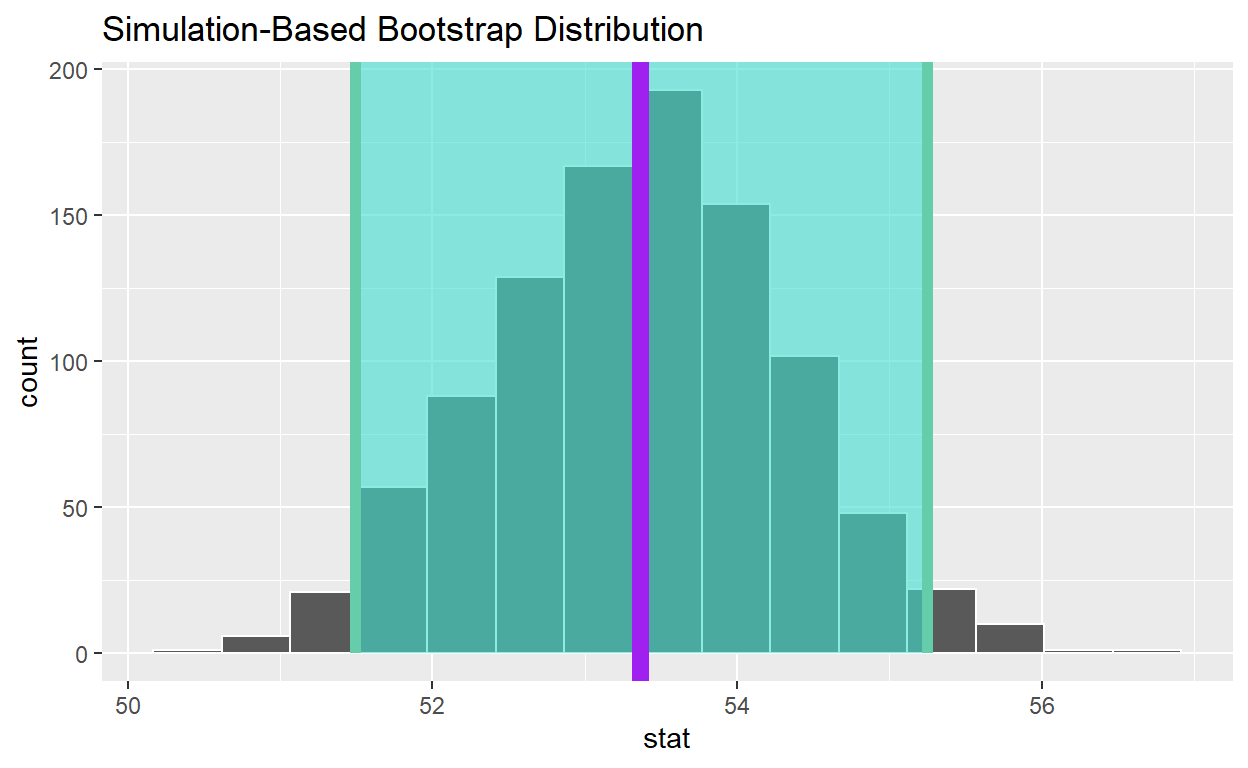
Calculate the population mean to see if it is in the 95% confidence interval
Assign the output to pop_mean_age
Display pop_mean_age
pop_mean_age <- congress_age_100 %>%
summarize(pop_mean= mean(age)) %>% pull()
pop_mean_age
[1] 53.36Add a line to the visualization at the, population mean, pop_mean_age, to the plot color it “purple”
visualize(bootstrap_distribution_mean_age) +
shade_confidence_interval(endpoints = congress_ci_percentile) +
geom_vline(xintercept = obs_mean_age, color = "hotpink", size = 1) +
geom_vline(xintercept = pop_mean_age , color = "purple", size = 3)
hr <- read_csv("https://estanny.com/static/week13/data/hr_3_tidy.csv",
col_types = "fddfff")
skim(hr)
| Name | hr |
| Number of rows | 500 |
| Number of columns | 6 |
| _______________________ | |
| Column type frequency: | |
| factor | 4 |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| gender | 0 | 1 | FALSE | 2 | fem: 253, mal: 247 |
| evaluation | 0 | 1 | FALSE | 4 | bad: 148, fai: 138, goo: 122, ver: 92 |
| salary | 0 | 1 | FALSE | 6 | lev: 98, lev: 87, lev: 87, lev: 86 |
| status | 0 | 1 | FALSE | 3 | fir: 196, pro: 172, ok: 132 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| age | 0 | 1 | 39.41 | 11.33 | 20 | 29.9 | 39.35 | 49.1 | 59.9 | ▇▇▇▇▆ |
| hours | 0 | 1 | 49.68 | 13.24 | 35 | 38.2 | 45.50 | 58.8 | 79.9 | ▇▃▃▂▂ |
null_t_distribution <- hr %>%
specify(response = age) %>%
hypothesize(null = "point", mu = 48) %>%
generate(reps = 1000, type = "bootstrap") %>%
calculate(stat = "mean")
null_t_distribution
# A tibble: 1,000 x 2
replicate stat
* <int> <dbl>
1 1 47.9
2 2 49.4
3 3 48.7
4 4 48.1
5 5 47.2
6 6 47.9
7 7 47.7
8 8 47.4
9 9 47.4
10 10 48.4
# ... with 990 more rowshr_2 <- read_csv("https://estanny.com/static/week13/data/hr_3_tidy.csv", col_types = "fddfff")
hr_2 %>%
group_by(gender) %>%
skim()
| Name | Piped data |
| Number of rows | 500 |
| Number of columns | 6 |
| _______________________ | |
| Column type frequency: | |
| factor | 3 |
| numeric | 2 |
| ________________________ | |
| Group variables | gender |
Variable type: factor
| skim_variable | gender | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|---|
| evaluation | male | 0 | 1 | FALSE | 4 | bad: 72, fai: 67, goo: 61, ver: 47 |
| evaluation | female | 0 | 1 | FALSE | 4 | bad: 76, fai: 71, goo: 61, ver: 45 |
| salary | male | 0 | 1 | FALSE | 6 | lev: 47, lev: 43, lev: 43, lev: 42 |
| salary | female | 0 | 1 | FALSE | 6 | lev: 51, lev: 46, lev: 45, lev: 43 |
| status | male | 0 | 1 | FALSE | 3 | fir: 98, pro: 81, ok: 68 |
| status | female | 0 | 1 | FALSE | 3 | fir: 98, pro: 91, ok: 64 |
Variable type: numeric
| skim_variable | gender | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|---|
| age | male | 0 | 1 | 38.23 | 10.86 | 20 | 28.9 | 37.9 | 47.05 | 59.9 | ▇▇▇▇▅ |
| age | female | 0 | 1 | 40.56 | 11.67 | 20 | 31.0 | 40.3 | 50.50 | 59.8 | ▆▆▇▆▇ |
| hours | male | 0 | 1 | 49.55 | 13.11 | 35 | 38.4 | 45.4 | 57.65 | 79.9 | ▇▃▂▂▂ |
| hours | female | 0 | 1 | 49.80 | 13.38 | 35 | 38.2 | 45.6 | 59.40 | 79.8 | ▇▂▃▂▂ |
hr_2 %>%
specify(response = hours, explanatory = gender) %>%
hypothesize(null = "independence") %>%
generate(reps=1000,type= "permute")
Response: hours (numeric)
Explanatory: gender (factor)
Null Hypothesis: independence
# A tibble: 500,000 x 3
# Groups: replicate [1,000]
hours gender replicate
<dbl> <fct> <int>
1 59.9 male 1
2 37 female 1
3 38.9 female 1
4 36.4 male 1
5 58.7 male 1
6 35 female 1
7 38.4 female 1
8 70.6 female 1
9 38.9 female 1
10 57 male 1
# ... with 499,990 more rows